Introduction
In the realms of functional genomics and epigenetics, the elucidation of transcription factor binding sites (TFBS) has perennially stood as a focal point of research endeavors. Disruption of TFBS has been intricately linked to phenotypic diversity, encompassing pivotal adaptive traits in agriculture and various disease states, including cancer. The comprehensive mapping of individual TFs' genome-wide binding profiles is paramount in deciphering the mechanisms underlying these changes. Traditional methods such as Chromatin Immunoprecipitation Sequencing (ChIP-seq), effective in detecting TFBS under optimal antibody quality, encounter limitations in non-model organism species where specific antibodies may not be readily available, thereby impeding the broader application of ChIP-seq. In 2016, O'Malley et al. published a seminal article in Cell utilizing DNA Affinity Purification Sequencing (DAP-seq) technology, swiftly delineating both the cis and trans regulatory landscapes of TFs targeting DNA regions. Within this work, they identified binding sites of 529 TFs in Arabidopsis thaliana, elucidating the impact of methylation status on the binding patterns of TFs. Subsequently, in 2017, Bartlett et al. detailed the experimental methodologies of DAP-seq in greater depth in Nature Protocols. The emergence of DAP-seq technology has liberated TFBS research from species constraints and antibody quality limitations, furnishing the fields of life sciences and medical research with a novel and potent tool for the investigation of TFs. How to choose ChIP-seq or DAP-seq, you can refer to "DAP-seq vs ChIP-seq".
DAP-seq emerges as a nascent genomic technology, amalgamating DNA affinity purification with high-throughput sequencing methodologies, aimed at elucidating the intricate interplay between DNA and specific binding proteins within the genome. Initially employed for the isolation and identification of specific DNA sequences interacting with proteins, DNA affinity purification techniques have evolved in tandem with the development of high-throughput sequencing technologies. DAP-seq has propelled this methodology to new heights, facilitating rapid and efficient analyses of the entire genome, thereby unveiling the intricate network of DNA-protein interactions inherent within the genome.
Principle of DAP-Seq
DAP-seq is a high-throughput method for screening TFBS, accomplished by first conducting affinity purification of genomic DNA bound by TFs exogenously expressed in vitro, followed by high-throughput sequencing to identify the TFBS. The principle underlying DAP-seq technology combines DNA affinity purification with high-throughput sequencing techniques. This methodology involves the exogenous expression of TFs containing an affinity tag, whereby the coding sequences of TFs are integrated into vectors containing the affinity tag (Halo-Tag), resulting in the formation of fusion proteins. Subsequently, these affinity-tagged TFs form complexes with DNA within the genome. The TF-DNA complexes are then extracted from cells or tissues using affinity purification methods. Subsequent high-throughput sequencing of the DNA allows for the identification of DNA sequences bound by TFs. Ultimately, analysis of the sequencing data enables the determination of TF binding positions within the genome, thereby generating genome-wide maps of TFBS.
A commonly used method for in vitro protein expression in DAP-seq is the Wheat Germ Cell-Free Protein Synthesis System (WGCF). Cell-free protein expression systems utilize exogenous DNA or mRNA templates in cell lysates along with a combination of cellular organelles and various enzymes, supplemented with amino acids, T7 RNA polymerase, and energy-regenerating substances, to express proteins outside of cells. These systems can utilize cell lysates from different sources, including Escherichia coli cells, wheat germ cells, rabbit reticulocytes, yeast cells, and insect cells. Compared to traditional in vivo cell-based protein expression systems, cell-free protein expression systems offer advantages such as rapid and precise protein expression, controllable protein expression and modification, and independence from cellular growth metabolism, ensuring reliable protein expression.
Workflow of DAP-Seq
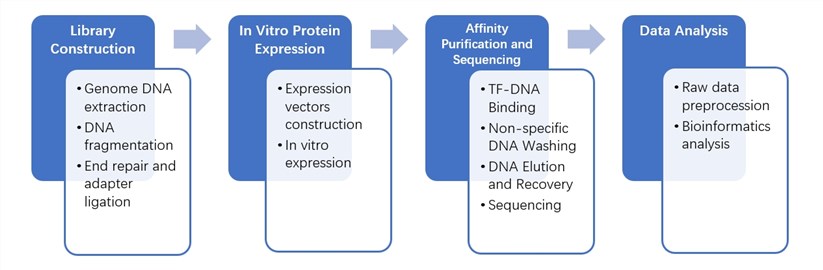
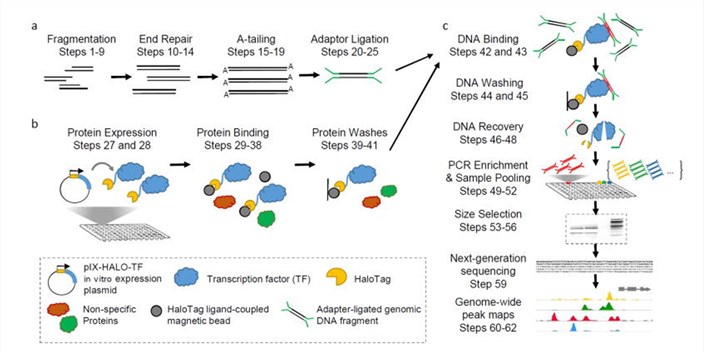 DAP-seq protocol overview(Bartlett et al., 2017)
DAP-seq protocol overview(Bartlett et al., 2017)
Applications of DAP-Seq
The DAP-seq technique is primarily employed to detect the binding sites of TFs or DNA-binding proteins across the genome, thereby facilitating the identification of downstream target genes and enhancing our understanding of TF functionality and regulatory mechanisms. The advent of DAP-seq has liberated TFBS research from species limitations and antibody quality constraints. This method finds application not only in model plant studies but also in investigations involving non-model plants, particularly within the realm of botanical research. Serving as a potent tool in plant epigenomics and chromatin accessibility studies, DAP-seq significantly broadens and deepens our comprehension of TFBS landscapes. Thus, it represents an innovative and efficient asset for advancing research in the field of life sciences, specifically pertaining to the exploration of TFs.
Identification of TFBSs
DAP-seq is instrumental in identifying TFBS, elucidating their pivotal regulatory roles in biological processes. For instance, researchers utilized DAP-seq technology to identify the binding motif and target genes of an NAC TF in the legume model plant Medicago truncatula, revealing the crucial regulatory role of NAC094 in nitrate-induced root nodule senescence. This discovery lays a significant foundation for breeding new crop varieties with efficient nitrogen utilization traits. Additionally, scientists employed DNA-seq to identify the binding motif and target genes of the PtoWRKY68 TF in Populus tomentosa. It unveiled the molecular mechanism by which genetic variations in the PtoWRKY68 gene regulate the response to drought stress through the ABA signaling pathway, providing a genetic basis for developing drought-tolerant tree varieties using molecular breeding strategies. These findings underscore the importance of DAP-seq in unraveling TF functions and regulatory networks, offering critical theoretical and practical support for genetic improvement and molecular breeding strategies in related fields.
Comparative Genomics of TF Binding
DAP-seq has been applied to compare the TF binding patterns across different plant species. For instance, researchers elucidated species-specific and conserved regulatory elements by comparing the TF binding patterns between Arabidopsis thaliana and maize, providing insights into the evolution and divergence of transcriptional regulatory networks.
Functional Characterization of TFs
DAP-seq can be utilized for studying TF functionality, thereby elucidating its regulatory networks. Researchers employed DAP-seq technology to identify the binding motif and target genes of the TF ONAC083 in rice. Further investigations revealed that OsNAC083 negatively regulates rice resistance to blast disease by binding to the ACGCAA element and modulating the transcription of OsRFPH2-6. This advancement enhances our understanding of the gene regulatory networks in rice and contributes to the development of crop improvement strategies.
For further insights into the applications of DAP-seq, please refer to the article"The Applications of DAP-seq Technology".
Advantages and Limitations of DAP-seq
The DAP-seq technique offers significant advantages in high throughput, antibody-independent operation, and wide applicability, enabling rapid and precise identification of TFBS, thereby expanding the scope of TF research. However, it demands substantial initial materials, entails complex data analysis, and lacks direct recognition of protein-DNA modifications. These factors may limit its applicability and interpretative capabilities under certain circumstances.
| Feature | DAP-seq | ChIP-seq |
| Experimental Mode | In vitro | In vivo |
| Antibody Dependence | No | Yes |
| Applicable to Non-model Plants | Yes | No |
| Time Cost | Low | High |
| High Throughput | Yes | No |
Conclusion and Prospectof DAP-seq
The DAP-seq technique stands as a powerful method for elucidating TFBS ex vivo, serving as a cornerstone in the realm of epigenetics. By employing this approach, researchers can swiftly and precisely identify the precise loci where TFs interact with genomic DNA, thereby unraveling the intricate functions and regulatory mechanisms embedded within gene regulatory networks. DAP-seq offers notable advantages, including its liberation from antibody constraints, broad applicability, and capacity for high throughput analysis, rendering it a favored avenue for probing into TFBS. However, the efficacy of DAP-seq is significantly contingent upon the nature of the TF, with success rates varying across distinct TF families. To date, researchers have harnessed DAP-seq technology to scrutinize the genomes of over 100 species.
Looking forward, amidst the continuum of technological advancements and refinements, DAP-seq holds even more expansive horizons for development. Firstly, as experimental methodologies evolve, we anticipate significant strides in the automation and quality control of DAP-seq protocols, promising augmented experimental efficiency and data fidelity. Secondly, propelled by the relentless progression of bioinformatics and computational technologies, DAP-seq data analysis methodologies will evolve to be more intricate and efficient, affording researchers deeper insights into the interpretation and functional elucidation of TFBS. Furthermore, through the integration with complementary omics technologies such as RNA-seq and ATAC-seq, DAP-seq harbors the potential to furnish a holistic comprehension of the intricate dynamics and complexities inherent within gene regulatory networks. Lastly, by refining the DAP-seq technique to enhance experimental precision and its adaptability to a wider array of binding site identifications, we pave the way for more robust and diversified applications.
In summation, the future trajectory of DAP-seq technology holds immense promise and propitious prospects within the domain of TF research. It is poised to provide us with more comprehensive and profound methodologies for delineating and dissecting TFBS, thereby catalyzing advancements and fostering progress within the realm of life sciences.
References
- O'Malley RC, Huang SC, Song L, et al. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape.Cell. 2016;165(5):1280-1292. doi:10.1016/j.cell.2016.04.038
- Bartlett A, O'Malley RC, Huang SC, et al. Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat Protoc. 2017;12(8):1659-1672. doi:10.1038/nprot.2017.055
- Wang L, Tian T, Liang J, et al. A transcription factor of the NAC family regulates nitrate-induced legume nodule senescence. New Phytol. 2023;238(5):2113-2129. doi:10.1111/nph.18896
- Fang Y, Wang D, Xiao L, et al. Allelic variation in transcription factor PtoWRKY68 contributes to drought tolerance in Populus. Plant Physiol. 2023;193(1):736-755. doi:10.1093/plphys/kiad315
- Bi Y, Wang H, Yuan X, et al. The NAC transcription factor ONAC083 negatively regulates rice immunity against Magnaporthe oryzae by directly activating transcription of the RING-H2 gene OsRFPH2-6. J Integr Plant Biol. 2023;65(3):854-875. doi:10.1111/jipb.13399